Abstract
Background: Therapy-related myeloid neoplasm (t-MN) is a clinicopathological entity that includes therapy-related myelodysplastic syndrome (t-MDS), therapy-related myelodysplastic syndrome/myeloproliferative neoplasm (t-MDS/MPN), and therapy-related acute myeloid leukemia (t-AML). Given that t-MN encompasses multiple phenotypes, an all-inclusive risk model for t-MN has not been proposed. Moreover, widely used risk stratification tools are limited to a single phenotype. For example, International prognostic scoring system (IPSS), revised-IPSS (IPSS-R) and World Health Organization based prognostic score (WPSS) for MDS and European LeukemiaNet classification for AML). Moreover, prognostic models such as IPSS, IPSS-R and WPSS did not include t-MN counterparts. Our aim was to develop and validate a risk stratification model that uses routinely clinical and cytogenetic factors available at t-MN diagnosis.
Methods: We retrospectively analyzed clinicopathological features and outcomes of 653 WHO-defined t-MN patients treated at Mayo Clinic, Rochester (US cohort, n=342) or South Australia MDS Registry (AU cohort; n=311) who had cytogenetics performed at t-MN diagnosis. Karyotypes were classified using the International System for Cytogenetic Nomenclature Criteria. Number of cytogenetic aberrancies were counted (Hasse et al, Leukemia 2019). A random forest survival algorithm was used to build prognostic model in the training cohort (US cohort). The model algorithm randomly bootstraps the original data into two thirds, where the model is developed, and one third, where the model is internally validated. The process is repeated multiple times to assure reproducibility of the final result. The model was subsequently validated in an independent AU cohort. The risk score optimal cut-off was determined using the maximally selected rank statistics. All the statistical analysis was performed using R statistical software.
Results: In the US cohort, the most common cytogenetic abnormalities were complex karyotype (CK, 49%), monosomal karyotype (MK, 46%), deletion 7q (del 7q, 36.5%) and deletion 5q (del 5q, 26.5%). The frequency of the most common cytogenetic abnormalities was different in the AU cohort: CK (26.8%), MK (26.2%), del 7q (25.2%) and del 5q (16.0%). Despite the difference, the median overall survival (OS) was not different between the US and AU cohorts (15 vs. 12 months, P =0.81).
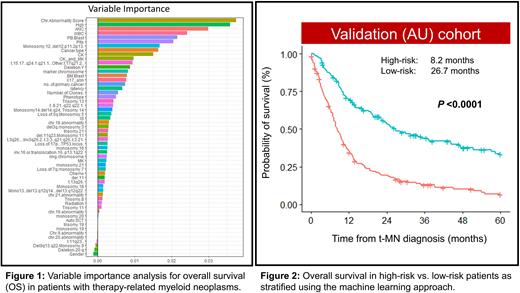
The US cohort was used to build the prognostic model and included 56 variables (15 clinical and 41 cytogenetic). Variables importance analysis shows the top variables from the most important to the least important that affect OS in the new model (Fig. 1). The variables with the highest impact on OS included the number of chromosomal abnormalities followed by blood counts, CK, and primary cancer type. In contrast, traditionally recognized high-risk features such as bone marrow blast percentage, chromosome 5 and/or 7 abnormalities had modest impact on OS in our model.
We next divided the training cohort into high- and low-risk groups based on the risk score derived from the model. In the US cohort, 208 (61%) and 134 (39%) patients were classified as high- and low-risk, respectively with a statistically significant difference in OS (8.42 vs. 40.03 months; P <0.001). The model was validated in the AU cohort. In the AU cohort, there were 174 (56%) patients with high risk and 137 (44%) patients with low risk. The model was very well-calibrated in the validation cohort, with significant median survival difference between the high-risk and low-risk group (8.2 vs. 26.7 months; P <0.001, Fig. 2). The concordance index of the model was 0.64 and 0.69 in the training model and the validation cohorts, respectively.
Conclusion: We developed and validated a prognostic model for all t-MN subtypes that incorporates routinely available information and is applicable to the day-to-day practice. We also identified novel factors including chromosomal abnormality number, CBC parameters, and the type of primary disease as those with the highest impact on survival. It is hoped that the incorporation of widely available and standardized testing that accurately identifies patients with extremely poor survival, will help prompt decision making-minimizing toxicities, including financial toxicities.
Disclosures
Sharplin:Novartis: Other: Travel and conference funding; Kite: Honoraria. Al-Kali:Astex: Other: research support to institution. Yeung:Takeda: Honoraria, Membership on an entity's Board of Directors or advisory committees; Amgen: Honoraria; Pfizer: Honoraria; BMS: Honoraria, Research Funding; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding. Litzow:Abbvie: Research Funding; Amgen: Research Funding; Astellas: Research Funding; Novartis: Research Funding; Syndax: Research Funding; Jazz: Consultancy; Actinium: Research Funding; Pluristem: Research Funding; Biosight: Other: Data Monitoring Board. Ross:Takeda: Membership on an entity's Board of Directors or advisory committees; Novartis: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Keros: Consultancy; Celgene: Research Funding; BMS: Honoraria. Mangaonkar:Bristol Myers Squibb: Research Funding. Patnaik:Kura Oncology, Stem Line Pharmaceuticals: Research Funding. Shanmuganathan:Amgen: Other: Meeting sponsorship; Novartis: Honoraria. Shah:Astellas: Research Funding; Celgene: Research Funding; Marker Therapeutics: Research Funding. Hiwase:BMS: Consultancy, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Novartis: Speakers Bureau; AbbVie: Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal